



BDBag API and CLI Source Code BDBag Source Code for GUI version
The BD Bag software allows researchers to address a significant Big Data challenge of assembling, identifying, and providing access to subsets of data in a large and complex data collection workflow such as from a catalog search to an analysis pipeline and to a publication service. This collection of utilities work with BagIt packages that conform to the BDDS Baggit and BDDS Bagit/RO profiles. A unique aspect of this work is that the data that is aggregated need not be collocated: instead, data collections can be uniquely identified where large elements may be located in cloud or enterprise storage. This is critical for big data elements where the cost of transfer of the data can be prohibitive. Another important feature is the use of JSON-LD to provide a standard way for linking metadata with existing ontologies and vocabularies. As the first example use of JSON-LD metadata, a model has been developed for representing ontology-based file types.
The BDBag GUI provides an intuitive graphical user interface for working with BDBags. Users can create and update bags, and also validate, archive, and fetch remote files. Binary executables for Windows and MacOS are provided, making it very easy for a user to get started working with BDBags.
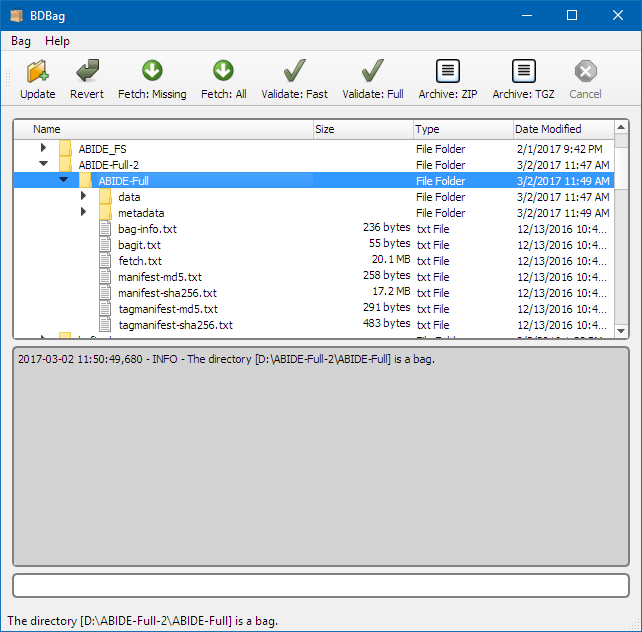
For more information and filing bug reports, see the project GitHub repository at http://github.com/ini-bdds/bdbag. Released versions of the software can be downloaded from https://github.com/ini-bdds/bdbag/releases.
BDBags have been used extensively in the following “Use Cases”: