



Get Latest BDDS Galaxy cloud profiler Software
Rapidly decreasing sequencing costs have led to a tremendous increase in the amount of sequence data to manage and analyze. For example, researchers who were previously limited to targeted exome sequencing in order to save costs can now apply whole genome sequencing to detect variants throughout the entire genome. The development of cost effective sequencing methods has also spurred an increase in the development of sequence analysis tools. For instance, researchers can now choose from among multiple DNA and RNA sequence aligners and variant callers. The explosion in both data and available analyses creates exciting opportunities for research advances but also presents challenges to researchers who lack adequate computing infrastructure and technical expertise. If technology expertise and large computer systems are required to reap the benefits of high throughput sequencing, then many researchers will be excluded from data-driven discovery, hindering the advance of biomedical science.
BDDS Galaxy addresses the challenges that researchers face when dealing with NGS analysis on a large scale. It combines state-of-the-art algorithms with sophisticated data management tools, a powerful graphical workflow environment, and a cloud-based elastic computational infrastructure. The following picture shows how BDDS Galaxy is leveraged for large scale analysis in conjunction with the rest of the BDDS tools.
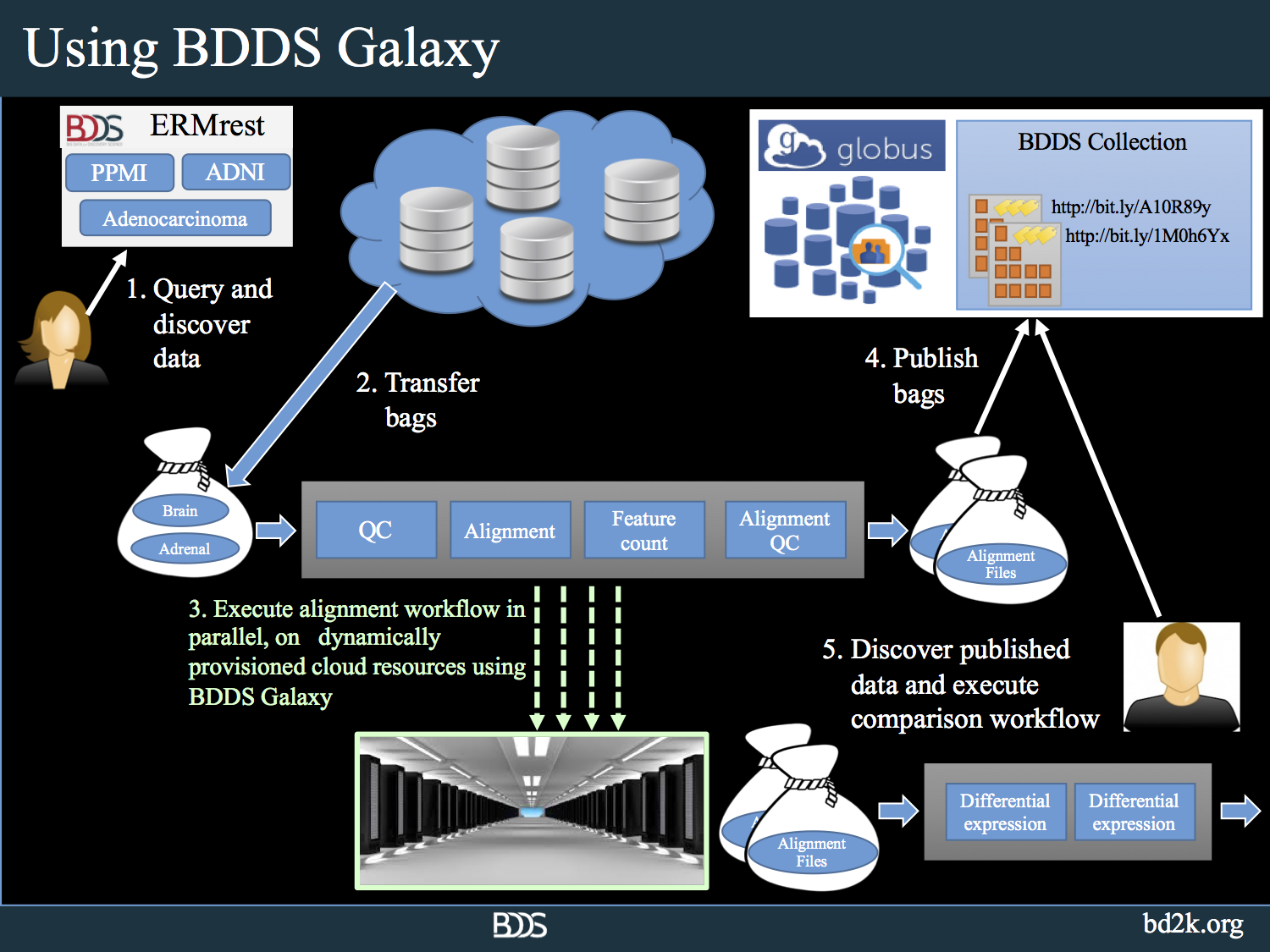
Using BDDS Galaxy, researchers can upload big biomedical genomics data to the tool using high speed, reliable Globus data movement services, interactively inspect the data using hundreds of NGS, proteomics analysis tools. The users can create workflows that would perform quality control and analyze datasets. Once the analysis protocol is stabilized, users can turn the created workflows into high performance workflows using batch submission tools that we developed.
In BDDS Galaxy we have implemented tools that would optimize the cost and performance of performing large-scale analytical workflows. We developed application profiling service and a cloud scheduler. The application profiling service generates computational profiles of analysis tools that the cloud scheduler leverages to perform large-scale analysis. These can be downloaded and used from here.
The following link provides a step-by-step guide to perform large-scale parallel analysis on Proteomics data using the TransProteomics Pipeline (TPP) Tutorial.
View information on BDDS Galaxy support for the BDBag and MINID toolset. View “Use Case” details for the following: